Direct contributions
Submitting a direct contribution is the fastest and easiest way to publish 3D surface models in M3 (and, as an option, CT data and MRI data as well). All you need is:
- A direct contribution manuscript file (which should remain as much concise as possible)
- A 3D data distribution statement
- One figure (representing one or several 3D models)
- At least one specimen with associated 3D surface data expressed in mm. enclosed in a .zip file containing one or more surface (.PLY or .VTP/.VTK) files. Blender projects (.BLEND) are also accepted.
About direct contributions
A direct contribution consists of a 3D dataset (up to 50 specimens) and of basic information regarding the concerned specimens and the 3D models.
The owners of a 3D dataset of specimens may want or
need to give public access to their data for different reasons:
- The 3D dataset has been already used and analyzed in another scientific journal.
- The analysis of this 3D dataset will soon be submitted for publication or is currently being reviewed in a scientific journal
which asks to give open access to the original data as soon as the article is published.
In that case, we ask you to keep the following timeline:
1) Submit your dataset to MorphoMuseuM. For each uploaded model, M3 provides you a temporary direct download link. You may transmit to third parties these temporary download links if you need to (some journals will require that you do so).
2) Once accepted for publication in MorphoMuseuM (it should only take a few days), we will provide you a citation DOI link related to your direct contribution.
3) Please insert a reference to this DOI inside your article (this should be possible during the revision process or during the proof edition process). For instance, you may use this DOI in your bibliography.
4) Once your article is published, unlock your dataset in MorphoMuseuM.
Note that your 3D Dataset will remain locked until your article is published. M3 will not give public access to your dataset until you inform us that the related article has been published. Also keep in mind that we may reject 3D datasets which have been locked for more than 6 months. - This 3D dataset may also not relate to any scientific publication, but the authors or the institution owning these data want to give public access to these models.
As stated above, M3 is the partner journal of Palaeovertebrata, in which we encourage you to publish your analyses of 3D datasets.
Direct contribution: guide for Authors
Please prepare the following documents before clicking on 'submit an article' button:
- text file of your Direct Contribution Manuscript (.doc, .docx, .odt, or .txt)*,
- A fully completed and signed 3D distribution statement (.doc, .docx, .odt or .pdf)*,
- At least one figure (.jpg, .tif, .eps, .pdf or .png) *
- As an option, you may wish to fill and attach the Specimen list table.
*: required for submissions
3D surface data:
Keep in mind that M3 primarily publishes "enriched" (= segmented, labelled, oriented, tagged or virtual restorations of damaged specimens) 3D surface models of vertebrate specimens.
- 3D data related to a given specimen must be compressed into one single .zip file.
- 3D surfaces should be expressed in millimeters (mm).
- 3D data format must be openable with freeware such as MeshLab, ImageJ, MorphoDig or other free software.
- Each .zip file should contain at least one surface file. ALL surfaces should be expressed in millimeters (mm). Please use .PLY or .VTP/.VTK formats for 3D surfaces enclosed in the .zip file (please convert .STL files as .PLY files using a freeware such as MeshLab). Point clouds are not considered as surfaces. Blender projects (.BLEND) are also accepted.
- Each .zip file should not contain any subfolder.
- Name the files enclosed inside the .zip file in a consistent and understandable manner: for instance, prefix all file names relative to one specimen with the inventory number of that specimen.
- Only include once the same piece of information inside the .zip file: for instance, avoid including the same 3D surface in different formats.
- If you have used MorphoDig to process your 3D surface(s), we encourage you to enclose as well position and orientation files (.pos and .ori ), .tag and .flg files (if applicable, see below) and project files (.ntw).
- Whether or not you have used MorphoDig to process your 3D surface(s), we ask you to provide your 3D models in a biologically relevant position and orientation. You can easily achieve this easily using MorphoDig. Non positioned/oriented 3D models might be rejected for publication.
- MorphoMuseuM displays color legends. If your surface models contain several distinct objects rendered with different colors, or if you have colored your 3D surface files,
we encourage you to enclose a .tag file inside your .zip file. You can produce a .tag file with MorphoDig or create it manually.
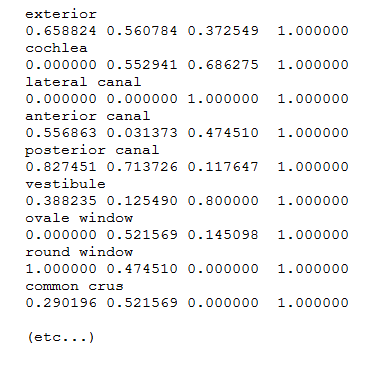
Tag file example
Tag files (.tag) are text files containing n pairs of lines, each pair being constructed following way:
line 2*n -1: name of the colored structure/object
line 2*n: corresponding color (red, green, blue) and transparency
- MorphoMuseuM displays flags, which are "clickable" structures of interest. If your plan to use this feature, we encourage you to enclose one or several .flg files inside your .zip file. You can produce .flg files with MorphoDig or create them manually.
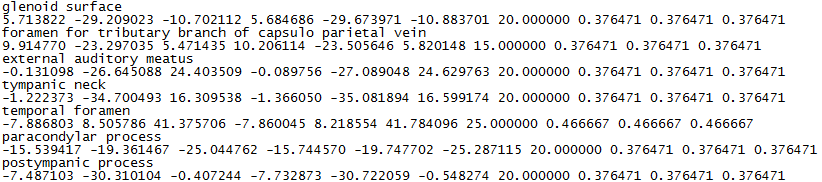
Flag file example
Flag files (.flg) are text files containing n pairs of lines, each pair being constructed following way:
line 2*n -1: name of the structure of interest
line 2*n: coordinates of the structure of interest (x, y, z), flag orientation (nx, ny, nz), flag length (in mm) and flag color (r, g, b).
If you have constructed manually your .flg file(s) and are not able to set flag orientation and length, you can set nx, ny, and nz to 0.0, and flag length to 1.0.
CT/MRI data:
M3 optionally publishes CT scan / MRI representations of vertebrate specimens. Such data must meet the following requirements:
- CT or MRI data related to a given specimen must be compressed into one single .zip file, weighing up to 126 Mo. If your original CT/MRI dataset weighs more than 126 Mo, you may read our image stack optimization page. If you still do not manage to create a file weighing less than 126 Mo, you may consider to create a Dryad dataset instead, and to inform us that it should be linked to your submission.
- We recommend to use .mhd, .mha or .raw format for CT/MRI Data. However, image stacks (.bmp, .tif, .dcm) are also accepted.
- Only include once the same piece of information inside the .zip file: for instance, avoid to include the same CT/MRI data in different formats or orientations.
File size limitation for CT/MRI files: 126 Mo/file
You may think that 126 Mo is ridiculously small for a zipped CT/MRI dataset, but then again, we encourage you to read our image stack optimization page.
Direct contribution: manuscript preparation
General recommendations:The manuscript of direct contributions should remain as much concise as possible.
Papers should conform to the following general layout:
Title page Section
The title page section must contain the following elements:- Title (it should be concise but informative, and where appropriate should include mention of family or higher taxon. Names of new taxa should not be given in title);
- Author's name (followed by a lower-case superscript letter immediately after the author's name and in front of the appropriate address);
- Authors' affiliation addresses (where the actual work was done) below the author's names (Institution's name, Department, city, state, ZIP code, and country);
- Corresponding author: Name, address, phone number, fax number, and e-mail address of the person who will handle correspondence at all stages of refereeing and publication, also post-publication. Only the email of the corresponding author is required.
- Abbreviated title (running headline) not to exceed 48 characters and spaces;
Abstract
This must be on a separate page.
- If the 3D dataset relates to a publication (published or under review), the abstract should refer explicitly to that publication. Abstract example:
The present 3D Dataset contains the 3D models analyzed in: Orliac M.J. 2012. The petrosal bone of extinct Suoidea (Mammalia, Artiodactyla). Journal of Systematic Palaeontology 11(8):925-945.
- If the 3D dataset does not relate to a publication, it should be about 100-250 words long and should summarize the contribution in a form that is intelligible in conjunction with the title. It should not include references.
For higher-rank taxonomic units use Latin names.
Keywords: The abstract should be followed by three to five keywords additional to those
in the title identifying the subject matter for retrieval systems. Keywords should
not exceed 85 characters and spaces.
Body of the manuscript
The manuscript has to be organized as follows:
- Introduction
- Methods
- Acknowledgements
- References
Introduction
All direct contributions published in M3 begin with a short introduction explaining the interest of the sample of 3D models. Authors may for instance briefly explain the purpose of the analysis published in the referred original article.
Methods
You manuscript should contain a brief description of the procedure(s)
used to produce the 3D surface models associated to your publication (e.g. manual or automatic segmentation...). Please also mention the software you have used.
Example of Methods section: The 3D surfaces were extracted semi-automatically within AVIZO 9.0 (Visualization Sciences Group) using the segmentation threshold selection tool.
The 3D surface models are provided in .ply format, and can therefore be opened with a wide range of freeware.
References
We recommend the use of a tool such as Endnote
or Mendeley for reference management and formatting. EndNote reference style for M3
can be downloaded here.
Mendeley reference style for M3
can be downloaded here.
In the text, give references in the following forms: "Sudre et al. (1989)", "Sudre (1974) said", "Sudre (1974: 28)"
where it is desired to refer to a specific page, and "(Sudre, 1983)" where giving reference
simply as authority for a statement. Note that names of joint authors are connected by '&'
in the text. The list of references must include all publications cited in the text and only these.
Prior to submission, make certain that all references in the text agree with those in the
references section, and that spelling is consistent throughout. In the list of references,
titles of periodicals must be given in full, not abbreviated. For books, give the title, place of
publication, name of publisher (if after 1930), and indication of edition if not the first.
In papers with half-tones, plate or figure citations are required only if they fall outside the
pagination of the reference cited.
References should conform as exactly as possible to one of these four styles, according
to the type of publication cited. When applicable, please include the DOI link associated to a given publication.
Agrasar E. L., 2004. Crocodilian remains from the
Upper Eocene of Dor-El-Talha, Libya. Annales de Paléontologie 90, 209-222. https://doi.org/10.1016/j.annpal.2004.05.001
McKenna M. C., Bell S. K., 1997. Classification of Mammals above the species level.
Columbia University Press, New York.
Shoshani J., West R. M., Court N., Savage R. J. G., Harris J. M., 1996. The earliest proboscideans: general plan, taxonomy and palaeoecology.
In: Shoshani J., Tassy P. (Eds.), The Proboscidea: Evolution and Palaeoecology of Elephants and
Relatives. Oxford Unioversity Press, Oxford, pp. 57-75.
Seiffert E. R., 2003. A phylogenetic analysis of living and extinct afrotherian placentals.
PhD, Duke University.
Other citations such as papers 'in press' [i.e. formally accepted
for publication] may appear on the list but not papers 'submitted' or 'in preparation'. These should
be cited as 'unpubl. data' in the text with the names and initials of all collaborators.
A personal communication may be cited in the text but not in the reference list. Please give all
surnames and initials for unpublished data or personal communication citations given in the text.
Tables
Keep these as simple as possible, with few horizontal and, preferably,
no vertical
rules. When assembling complex tables and data matrices, bear the dimensions of the printed page
(250 x 184.6 mm) in mind; reducing type-size to accommodate a multiplicity of columns will affect
legibility. Each table should be uploaded as a separate file.
Illustrations (optional)
As an option, you may include illustrations in your manuscript.
These normally include photographs, drawings, and diagrams in either black and white or color
(free of charge).
Use one consecutive set of Arabic numbers for all illustrations (do not separate 'Plates'
and 'Text-figures' - treat all as 'Figures'). Figures should be numbered in the order in which they
are cited in the text. Use upper case letters for subdivisions (e.g. Figure 1A-D) of figures; all
other lettering should be lower case. All lettering should be in Arial font. Required resolution: 300 dpi at the final required image
size. The labelling and any line drawings in a composite figure should be added in vector format.
Figures should not be embedded in the manuscript file; each figure should be uploaded as a separate
file.
Authors wishing to use illustrations already published must obtain written permission from
the copyright holder before submitting the manuscript.
Grouping and mounting: when
grouping photographs, aim to make the dimensions of the group (including guttering of 2 mm between
each picture) as close as possible to the page dimensions of 184.6 x 250 mm (for a two-column figure)
or 88.3 x 250 (for a one-column figure), thereby optimizing use of the available space. Remember that
grouping photographs of varied contrast can result in poor reproduction.









