Explodable 3D Dog Skull for Veterinary Education
3D model of Burmesescorpiops wunpawng
3D models of Miocene vertebrates from Tavers
3D GM dataset of bird skeletal variation
Skeletal embryonic development in the catshark
Bony connexions of the petrosal bone of extant hippos
bony labyrinth (11) , inner ear (10) , Eocene (8) , South America (8) , Paleobiogeography (7) , skull (7) , phylogeny (6)
Lionel Hautier (23) , Maëva Judith Orliac (21) , Laurent Marivaux (16) , Rodolphe Tabuce (14) , Bastien Mennecart (13) , Pierre-Olivier Antoine (12) , Renaud Lebrun (11)
|
3D models related to the publication: The ossicular chain of Cainotheriidae (Mammalia, Artiodactyla)
|

|
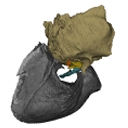
M3#508reconstruction of the middle ear with petrosal, bulla, stapes, incus, malleus Type: "3D_surfaces"doi: 10.18563/m3.sf.508 state:published |
Download 3D surface file |
This contribution contains the 3D model of an endocranial cast analyzed in “A 10 ka intentionally deformed human skull from Northeast Asia”. There are many studies on the morphological characteristics of intentional cranial deformation (ICD), but few related 3D models were published. Here, we present the surface model of an intentionally deformed 10 ka human cranium for further research on ICD practice. The 3D model of the endocranial cast of this ICD cranium was discovered near Harbin City, Province Heilongjiang, Northeast China. The fossil preserved only the frontal, parietal, and occipital bones. To complete the endocast model of the specimen, we printed a 3D model and used modeling clay to reconstruct the missing part based on the general form of the modern human endocast morphology.
Homo sapiens IVPP-PA1616 View specimen

|
M3#972The frontal region of the endocast is flattened, probably formed by the constant pressure on the frontal bone during growth. There is a well-developed frontal crest on the endocranial surface. The endocast widens posteriorly from the frontal lobe. The widest point of the endocast is at the lateral border of the parietal lobe. The lower parietal areas display a marked lateral expansion. The overall shape of the endocast is asymmetrical, with the left side of the parietal lobe being more laterally expanded than the right side. Like the frontal lobe, the occipital lobe is also anteroposteriorly flattened. Type: "3D_surfaces"doi: 10.18563/m3.sf.972 state:published |
Download 3D surface file |

|
M3#976The original endocranial cast model (with texture) of IVPP-PA1616. It shows the original structures of the specimen, and was not altered in any way. Type: "3D_surfaces"doi: 10.18563/m3.sf.976 state:published |
Download 3D surface file |
The present 3D Dataset contains the 3D model of a left dentary with m1-m3 analyzed in “A new fossil of Tayassuidae (Mammalia: Certartiodactyla) from the Pleistocene of northern Brazil”. The 3D model was generated using a laser scanning.
cf. Pecari tajacu UFSM 11606 View specimen

|
M3#498Left dentary with m1-m3 Type: "3D_surfaces"doi: 10.18563/m3.sf.498 state:published |
Download 3D surface file |

The present 3D Dataset contains the 3D models analyzed in: Perrichon et al. 2023. Ontogenetic variability of the intertympanic sinus distinguishes lineages within Crocodylia.
Mecistops sp. ag SVSTUA 022001 View specimen

|
M3#980Intertympanic sinus system (expressed in meters) Type: "3D_surfaces"doi: 10.18563/m3.sf.980 state:published |
Download 3D surface file |
Crocodylus niloticus ag SVSTUA 022002 View specimen

|
M3#981Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.981 state:published |
Download 3D surface file |
Mecistops sp. AMU Zoo 04721 View specimen

|
M3#982Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.982 state:published |
Download 3D surface file |
Crocodylus sp. MHNL QV14 View specimen

|
M3#983Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.983 state:published |
Download 3D surface file |
Crocodylus rhombifer MHNL 42006506 View specimen

|
M3#1008Intertympanic sinus system (incomplete) Type: "3D_surfaces"doi: 10.18563/m3.sf.1008 state:published |
Download 3D surface file |
Crocodylus rhombifer MHNL 42006507 View specimen

|
M3#1007Intertympanic sinus system (incomplete) Type: "3D_surfaces"doi: 10.18563/m3.sf.1007 state:published |
Download 3D surface file |
Crocodylus niloticus MHNL 50001387 View specimen

|
M3#1006Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1006 state:published |
Download 3D surface file |
Crocodylus palustris MHNL 50001388 View specimen

|
M3#1004Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1004 state:published |
Download 3D surface file |
Crocodylus porosus MHNL 50001389 View specimen

|
M3#1009Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1009 state:published |
Download 3D surface file |
Mecistops sp. MHNL 50001393 View specimen

|
M3#1010Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1010 state:published |
Download 3D surface file |
Crocodylus niloticus MHNL 50001397 View specimen

|
M3#1018Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1018 state:published |
Download 3D surface file |
Crocodylus porosus MHNL 50001398 View specimen

|
M3#1017Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1017 state:published |
Download 3D surface file |
Crocodylus niloticus MHNL 50001405 View specimen

|
M3#1016Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1016 state:published |
Download 3D surface file |
Gavialis gangeticus MHNL 50001407 View specimen

|
M3#1015Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1015 state:published |
Download 3D surface file |
Crocodylus niloticus MHNL 90001850 View specimen

|
M3#1014Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1014 state:published |
Download 3D surface file |
Crocodylus niloticus MHNL 90001851 View specimen

|
M3#1013Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1013 state:published |
Download 3D surface file |
Crocodylus niloticus MHNL 90001855 View specimen

|
M3#1012Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1012 state:published |
Download 3D surface file |
Osteolaemus tetraspis MHNM.9095.0 View specimen

|
M3#1011Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1011 state:published |
Download 3D surface file |
Voay robustus MNHN F.1908-5 View specimen

|
M3#1003Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1003 state:published |
Download 3D surface file |
Crocodylus sp. MNHN-F.1908-5-2 View specimen

|
M3#1005Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1005 state:published |
Download 3D surface file |
Osteolaemus tetraspis MZS Cro 040 View specimen

|
M3#1002Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1002 state:published |
Download 3D surface file |
Crocodylus acutus MZS Cro 055 View specimen

|
M3#991Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.991 state:published |
Download 3D surface file |
Melanosuchus niger MZS Cro 073 View specimen

|
M3#989Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.989 state:published |
Download 3D surface file |
Mecistops sp. MZS Cro 083 View specimen

|
M3#990Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.990 state:published |
Download 3D surface file |
Tomistoma schlegelii MZS Cro 094 View specimen

|
M3#988Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.988 state:published |
Download 3D surface file |
Gavialis gangeticus NHMUK 1846.1.7.3 View specimen

|
M3#987Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.987 state:published |
Download 3D surface file |
Osteolaemus tetraspis NHMUK 1862.6.30.5 View specimen

|
M3#986Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.986 state:published |
Download 3D surface file |
Gavialis gangeticus NHMUK 1873 View specimen

|
M3#985intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.985 state:published |
Download 3D surface file |
Tomistoma schlegelii NHMUK 1893.3.6.14 View specimen

|
M3#984Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.984 state:published |
Download 3D surface file |
Mecistops sp. NHMUK 1924.5.10.1 View specimen

|
M3#992Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.992 state:published |
Download 3D surface file |
Voay robustus NHMUK PV R 36684 View specimen

|
M3#993Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.993 state:published |
Download 3D surface file |
Voay robustus NHMUK PV R 36685 View specimen

|
M3#1001Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1001 state:published |
Download 3D surface file |
Crocodylus niloticus UCBL FSL 532077 View specimen

|
M3#1000Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1000 state:published |
Download 3D surface file |
Crocodylus porosus/siamensis UCBLZ 2019-1-237 View specimen

|
M3#999Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.999 state:published |
Download 3D surface file |
Osteolaemus tetraspis UCBLZ 2019-1-236 View specimen

|
M3#998Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.998 state:published |
Download 3D surface file |
Alligator mississipiensis UCBLZ WB35 View specimen

|
M3#997Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.997 state:published |
Download 3D surface file |
Crocodylus siamensis UCBLZ WB41 View specimen

|
M3#996Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.996 state:published |
Download 3D surface file |
Tomistoma schlegelii UM 1097 View specimen

|
M3#995Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.995 state:published |
Download 3D surface file |
Crocodylus niloticus UM 2001-1756-1-434 NR View specimen

|
M3#994Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.994 state:published |
Download 3D surface file |
Mecistops sp. UM N89 View specimen

|
M3#1019Intertympanic sinus system Type: "3D_surfaces"doi: 10.18563/m3.sf.1019 state:published |
Download 3D surface file |
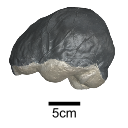
This contribution provides for the first time the 3D model of the type specimen of Molassitherium delemontense (Mammalia, Rhinocerotidae) described in the following publication: Becker et al. (2013), Journal of Systematic Palaeontology, Vol. 11, Issue 8, 947–972, https://doi.org/10.1080/14772019.2012.699007. Conservation issues of the specimen and solutions using 3D model and 3D prints are detailed.
Molassitherium delemontense MJSN POI007–245 View specimen

|
M3#384Skull of Molassitherium delemontense Becker and Antoine, 2013 (in Becker et al. 2013): holotype Type: "3D_surfaces"doi: 10.18563/m3.sf.384 state:published |
Download 3D surface file |
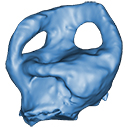
The present 3D Dataset contains the 3D models analyzed in "Neenan, J. M., Reich, T., Evers, S., Druckenmiller, P. S., Voeten, D. F. A. E., Choiniere, J. N., Barrett, P. M., Pierce, S. E. and Benson, R. B. J. Evolution of the sauropterygian labyrinth with increasingly pelagic lifestyles. Current Biology, 27." https://doi.org/10.1016/j.cub.2017.10.069
Amblyrhynchus cristatus OUMNH 11616 View specimen
|
M3#322Right labyrinth of Amblyrhynchus cristatus (OUMNH 11616). Type: "3D_surfaces"doi: 10.18563/m3.sf.322 state:published |
Download 3D surface file |
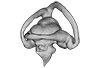
Augustasaurus hagdorni FMNH PR 1974 View specimen
|
M3#333Right labyrinth model of Augustasaurus FMNH PR 1974 Type: "3D_surfaces"doi: 10.18563/m3.sf.333 state:published |
Download 3D surface file |
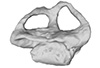
Callawayasaurus colombiensis UCMP V-38349 / UCMP V-125328 View specimen

|
M3#331Composite left labyrinth of Callawayasaurus. The majority of the model is from the holotype (UCMP V-38349), but the anterior portion is formed from the right labyrinth (reflected) from the paratype (UCMP V-125328). Type: "3D_surfaces"doi: 10.18563/m3.sf.331 state:published |
Download 3D surface file |
Lepidochelys olivacea SMNS 11070 View specimen
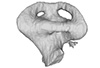
|
M3#330Left labyrinth model of Lepidochelys SMNS 11070 Type: "3D_surfaces"doi: 10.18563/m3.sf.330 state:published |
Download 3D surface file |
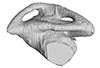
Macrochelys temminckii FMNH 22111 View specimen
|
M3#334Left labyrinth model of Macrochelys FMNH 22111 Type: "3D_surfaces"doi: 10.18563/m3.sf.334 state:published |
Download 3D surface file |
Macroplata tenuiceps NHMUK R 5488 View specimen

|
M3#328Left labyrinth of Macroplata NHMUK R 5488 Type: "3D_surfaces"doi: 10.18563/m3.sf.328 state:published |
Download 3D surface file |
Microcleidus homalospondylus NHMUK 36184 View specimen

|
M3#327Right labyrinth model of Microcleidus NHMUK 36184 Type: "3D_surfaces"doi: 10.18563/m3.sf.327 state:published |
Download 3D surface file |
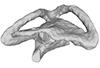
Nothosaurus sp. NME 16/4 View specimen
|
M3#326Right labyrinth model of Nothosaurus sp. NME 16/4 Type: "3D_surfaces"doi: 10.18563/m3.sf.326 state:published |
Download 3D surface file |
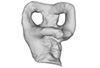
Peloneustes philarchus NHMUK R 3803 View specimen
|
M3#325Left labyrinth model of Peloneustes philarchus NHMUK R 3803 Type: "3D_surfaces"doi: 10.18563/m3.sf.325 state:published |
Download 3D surface file |
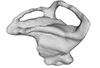
Placodus gigas UMO BT 13 View specimen
|
M3#324Right labyrinth model of Placodus gigas UMO BT 13 Type: "3D_surfaces"doi: 10.18563/m3.sf.324 state:published |
Download 3D surface file |
Puppigerus camperi NHMUK R 38955 View specimen

|
M3#323Left labyrinth model of Puppigerus NHMUK R 38955 Type: "3D_surfaces"doi: 10.18563/m3.sf.323 state:published |
Download 3D surface file |
Simosaurus gaillardoti GPIT RE/09313 View specimen
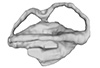
|
M3#332Right labyrinth model of Simosaurus GPIT RE/09313 Type: "3D_surfaces"doi: 10.18563/m3.sf.332 state:published |
Download 3D surface file |
Libonectes morgani SMUSMP 69120 View specimen

|
M3#335Right labyrinth model of Libonected morgani (SMUSMP 69120) Type: "3D_surfaces"doi: 10.18563/m3.sf.335 state:published |
Download 3D surface file |

This contribution contains the 3D models described and figured in: The Neogene record of northern South American native ungulates. Smithsonian Contributions to Paleobiology. Doi: 10.5479/si.1943-6688.101
Hilarcotherium miyou IGMp 881327 View specimen

|
M3#318Right upper M2 Type: "3D_surfaces"doi: 10.18563/m3.sf.318 state:published |
Download 3D surface file |
Hilarcotherium miyou MUN-STRI 34216 View specimen

|
M3#319Right upper P4 Type: "3D_surfaces"doi: 10.18563/m3.sf.319 state:published |
Download 3D surface file |

|
M3#320Right upper M2 Type: "3D_surfaces"doi: 10.18563/m3.sf.320 state:published |
Download 3D surface file |
Falcontoxodon aguilerai AMU-CURS 585 View specimen

|
M3#321Maxilla with left M3-P2 and right I2 Type: "3D_surfaces"doi: 10.18563/m3.sf.321 state:published |
Download 3D surface file |
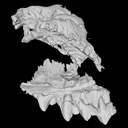
The present 3D Dataset contains the 3D model analyzed in the following publication: Solé et al. (2018), Niche partitioning of the European carnivorous mammals during the paleogene. Palaios. https://doi.org/10.2110/palo.2018.022
Hyaenodon leptorhynchus FSL848325 View specimen

|
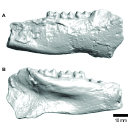
M3#336The specimen FSL848325 is separated in two fragments: the anterior part bears the incisors, the deciduous and permanent canines, while the posterior part bears the right P3, P4, M1 and M2. The P2 is isolated. When combined, the cranium length is approximatively 10.5 cm long. The anterior part is 6.9 cm long and 2.15 cm wide (taken at the level of the P1). The posterior part is 4.8 cm long. The anterior part of the cranium is very narrow. Type: "3D_surfaces"doi: 10.18563/m3.sf.336 state:published |
Download 3D surface file |
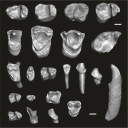
This contribution contains the three-dimensional digital models of the dental fossil material of anthropoid and strepsirrhine primates, discovered in Lower Oligocene detrital deposits outcropping in the Porto Rico and El Argoub areas, east of the Dakhla peninsula region (Atlantic Sahara; in the south of Morocco, near the northern border of Mauritania). These fossils were described, figured and discussed in the following publication: Marivaux et al. (2024), A new primate community from the earliest Oligocene of the Atlantic margin of Northwest Africa: Systematic, paleobiogeographic and paleoenvironmental implications. Journal of Human Evolution. https://doi.org/10.1016/j.jhevol.2024.103548
Catopithecus aff. browni DAK-Arg-087 View specimen

|
M3#1211Isolated right lower m3 (worn) Type: "3D_surfaces"doi: 10.18563/m3.sf.1211 state:published |
Download 3D surface file |
Catopithecus aff. browni DAK-Arg-088 View specimen

|
M3#1212Isolated right lower m2 (abraded/corroded) Type: "3D_surfaces"doi: 10.18563/m3.sf.1212 state:published |
Download 3D surface file |
Catopithecus aff. browni DAK-Arg-089 View specimen

|
M3#1213Isolated left lower m1 (worn) Type: "3D_surfaces"doi: 10.18563/m3.sf.1213 state:published |
Download 3D surface file |
Catopithecus aff. browni DAK-Pto-052 View specimen

|
M3#1214Isolated right lower m1 (pristine but lacking the mesiobuccal region) Type: "3D_surfaces"doi: 10.18563/m3.sf.1214 state:published |
Download 3D surface file |
Catopithecus aff. browni DAK-Arg-090 View specimen

|
M3#1215Isolated left upper P4 Type: "3D_surfaces"doi: 10.18563/m3.sf.1215 state:published |
Download 3D surface file |
Catopithecus aff. browni DAK-Arg-091 View specimen

|
M3#1216Isolated left upper M2 (worn and corroded) Type: "3D_surfaces"doi: 10.18563/m3.sf.1216 state:published |
Download 3D surface file |
Catopithecus aff. browni DAK-Pto-053 View specimen

|
M3#1217Isolated right upper M1 (lacking the buccal region) Type: "3D_surfaces"doi: 10.18563/m3.sf.1217 state:published |
Download 3D surface file |
Abuqatrania cf. basiodontos DAK-Arg-092 View specimen

|
M3#1218Isolated left lower c1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1218 state:published |
Download 3D surface file |
?Propliopithecus sp. DAK-Pto-056 View specimen

|
M3#1219Isolated right lower m3 (fragment of talonid of a germ) Type: "3D_surfaces"doi: 10.18563/m3.sf.1219 state:published |
Download 3D surface file |
Abuqatrania cf. basiodontos DAK-Arg-093 View specimen

|
M3#1469Isolated right lower m1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1469 state:published |
Download 3D surface file |
Abuqatrania cf. basiodontos DAK-Arg-094 View specimen

|
M3#1221Isolated left upper M1 or M2 (corroded, lacking the enamel cap [exposed dentine]) Type: "3D_surfaces"doi: 10.18563/m3.sf.1221 state:published |
Download 3D surface file |
Abuqatrania cf. basiodontos DAK-Arg-095 View specimen

|
M3#1222Isolated right lower i1 or i2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1222 state:published |
Download 3D surface file |
Abuqatrania cf. basiodontos DAK-Arg-096 View specimen

|
M3#1223Isolated right lower p2 (worn apex) Type: "3D_surfaces"doi: 10.18563/m3.sf.1223 state:published |
Download 3D surface file |
Abuqatrania cf. basiodontos DAK-Arg-097 View specimen

|
M3#1224Isolated right lower p2 (worn apex and broken root) Type: "3D_surfaces"doi: 10.18563/m3.sf.1224 state:published |
Download 3D surface file |
Afrotarsius sp. DAK-Arg-098 View specimen

|
M3#1225Isolated left lower p3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1225 state:published |
Download 3D surface file |
Afrotarsius sp. DAK-Pto-054 View specimen

|
M3#1226Isolated right lower m1 (abraded/corroded) Type: "3D_surfaces"doi: 10.18563/m3.sf.1226 state:published |
Download 3D surface file |
Orolemur mermozi DAK-Pto-055 View specimen

|
M3#1227Isolated right upper M1 or M2 (pristine, Holotype) Type: "3D_surfaces"doi: 10.18563/m3.sf.1227 state:published |
Download 3D surface file |
Wadilemur cf. elegans DAK-Arg-099 View specimen

|
M3#1228Isolated right lower m2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1228 state:published |
Download 3D surface file |
cf. 'Anchomomys' milleri DAK-Arg-100 View specimen

|
M3#1229Isolated right lower c1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1229 state:published |
Download 3D surface file |
Abuqatrania cf. basiodontos DAK-Arg-101 View specimen

|
M3#1396Isolated left upper M3 (abraded) Type: "3D_surfaces"doi: 10.18563/m3.sf.1396 state:published |
Download 3D surface file |
Orogalago saintexuperyi DAK-Arg-102 View specimen

|
M3#1397Isolated left lower m2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1397 state:published |
Download 3D surface file |
Wadilemur cf. elegans DAK-Arg-103 View specimen

|
M3#1473Isolated right upper M1 or M2 (lacking the mesial and buccal regions) Type: "3D_surfaces"doi: 10.18563/m3.sf.1473 state:published |
Download 3D surface file |

The present 3D Dataset contains the 3D models illustrated and described in the chapter “Paleoneurology of Artiodactyla, an overview of the evolution of the artiodactyl brain” (Orliac et al. 2022) published in "Paleoneurology of amniotes: new directions in the study of fossil endocasts", edited by Dozo, Paulina-Carabajal, Macrini and Walsh.
Homacodon vagans AMNH 12695 View specimen
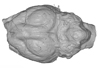
|
M3#1063Endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1063 state:published |
Download 3D surface file |
Helohyus sp. AMNH 13079 View specimen

|
M3#1064Endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1064 state:published |
Download 3D surface file |
Leptauchenia sp. AMNH 45508 View specimen

|
M3#1065endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1065 state:published |
Download 3D surface file |
Agriochoerus sp. AMNH 95330 View specimen

|
M3#1067endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1067 state:published |
Download 3D surface file |
Mouillacitherium elegans UM ACQ 6625 View specimen

|
M3#1068endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1068 state:published |
Download 3D surface file |
Caenomeryx filholi UM PDS 2570 View specimen

|
M3#1069endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1069 state:published |
Download 3D surface file |
Dichobune leporina MNHN.F.QU16586 View specimen

|
M3#1070endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1070 state:published |
Download 3D surface file |
Anoplotherium sp. not numbered View specimen

|
M3#1071endocranial cast Type: "3D_surfaces"doi: 10.18563/m3.sf.1071 state:published |
Download 3D surface file |
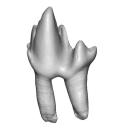
This contribution contains the 3D model described and figured in the following publication: Martin, T., Averianov, A. O., Schultz, J. A., & Schwermann, A. H. (2023). A stem therian mammal from the Lower Cretaceous of Germany. Journal of Vertebrate Paleontology, e2224848.
Spelaeomolitor speratus WMNM P99101 View specimen

|
M3#12573D_model_Spelaeomolitor_lower_molar Type: "3D_surfaces"doi: 10.18563/m3.sf.1257 state:published |
Download 3D surface file |

|
M3#1258CT imagestack (jpgs) and info data sheet (pca file) in one zip folder Type: "3D_CT"doi: 10.18563/m3.sf.1258 state:published |
Download CT data |

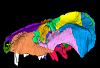
The present 3D Dataset contains the 3D model analyzed in the following publication: Carolina A. Hoffmann, A. G. Martinelli & M. B. Andrade. 2023. Anatomy of the holotype of “Probelesodon” kitchingi revisited, a chiniquodontid cynodont (Synapsida, Probainognathia) from the early Late Triassic of southern Brazil, Journal of Paleontology
Probelesodon kitchingi MCP 1600 PV View specimen
|
M3#11513D models of the skull with segmented bones and without the segmentation. colormap and orientation files also added. Type: "3D_surfaces"doi: 10.18563/m3.sf.1151 state:published |
Download 3D surface file |

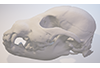
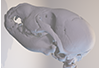
This contribution comprises the 3D models of three wolf pup skulls, which were used for the publication by Geiger et al. 2017 on Neomorphosis and heterochrony of skull shape in dog domestication.
Canis lupus CLL2 View specimen
|
M3#3123d model of a wolf pup skull Type: "3D_surfaces"doi: 10.18563/m3.sf.312 state:published |
Download 3D surface file |
Canis lupus CLL4 View specimen
|
M3#3133d model of a wolf pup skull Type: "3D_surfaces"doi: 10.18563/m3.sf.313 state:published |
Download 3D surface file |
Canis lupus CLL5 View specimen

|
M3#3143d model of a wolf pup skull Type: "3D_surfaces"doi: 10.18563/m3.sf.314 state:published |
Download 3D surface file |
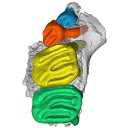
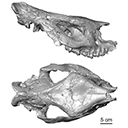
This contribution contains the 3D model of the holotype of Simplomys hugi, the new dormouse species from the locality of Glovelier described and figured in the following publication: New data on the Miocene dormouse Simplomys García-Paredes, 2009 from the peri-alpin basins of Switzerland and Germany: palaeodiversity of a rare genus in Central Europe. https://doi.org/10.1007/s12549-018-0339-y
Simplomys hugi MJSN-GLM017-0001 View specimen

|
M3#385the left maxilla with four teeth ( DP4, P4, M1 and M2) Type: "3D_surfaces"doi: 10.18563/m3.sf.385 state:published |
Download 3D surface file |
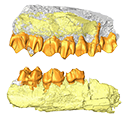
The present 3D Dataset contains 3D models of the holotypes described in Aiglstorfer et al. (2023a). Miocene Moschidae (Mammalia, Ruminantia) from the Linxia Basin (China) connect Europe and Asia and show early evolutionary diversity of a today monogeneric family. Palaeogeography, Palaeoclimatology, Palaeoecology.
Micromeryx? caoi CUGB GV 87045 View specimen

|
M3#11123D models of the holotype of “Micromeryx” caoi (CUGB GV87045) including the models of the teeth, the mandibule, and the sediment. Type: "3D_surfaces"doi: 10.18563/m3.sf.1112 state:published |
Download 3D surface file |
Hispanomeryx linxiaensis IVPP V28596 View specimen

|
M3#11133D models of the holotype of Hispanomeryx linxiaensis (IVPP V28596) including the models of the teeth, the mandibule, and the sediment. Type: "3D_surfaces"doi: 10.18563/m3.sf.1113 state:published |
Download 3D surface file |

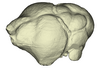
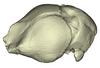
The present 3D Dataset contains the 3D models of extant Chiropteran endocranial casts, documenting 16 of the 19 extant bat families. They are used by Maugoust & Orliac (2023) to assess the correspondences between the brain and brain-surrounding tissues (i.e., neural tissues, blood vessels, meninges) and their imprint on the braincase, allowing for eventually proposing a Chiroptera-scale nomenclature of the endocast.
Balantiopteryx plicata UMMZ 102659 View specimen
|
M3#1132Endocranial cast of the corresponding cranium of Balantiopteryx plicata Type: "3D_surfaces"doi: 10.18563/m3.sf.1132 state:published |
Download 3D surface file |
Idiurus macrotis AMNH M-187705 View specimen
|
M3#1133Endocranial cast of the corresponding cranium of Nycteris macrotis Type: "3D_surfaces"doi: 10.18563/m3.sf.1133 state:published |
Download 3D surface file |
Thyroptera tricolor UMMZ 53240 View specimen

|
M3#1134Endocranial cast of the corresponding cranium of Thyroptera tricolor Type: "3D_surfaces"doi: 10.18563/m3.sf.1134 state:published |
Download 3D surface file |
Noctilio albiventris UMMZ 105827 View specimen

|
M3#1135Endocranial cast of the corresponding cranium of Noctilio albiventris Type: "3D_surfaces"doi: 10.18563/m3.sf.1135 state:published |
Download 3D surface file |
Mormoops blainvillii AMNH M-271513 View specimen

|
M3#1136Endocranial cast of the corresponding cranium of Mormoops blainvillii Type: "3D_surfaces"doi: 10.18563/m3.sf.1136 state:published |
Download 3D surface file |
Macrotus waterhousii UMMZ 95718 View specimen
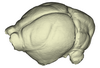
|
M3#1137Endocranial cast of the corresponding cranium of Macrotus waterhousii Type: "3D_surfaces"doi: 10.18563/m3.sf.1137 state:published |
Download 3D surface file |
Nyctiellus lepidus UMMZ 105767 View specimen
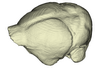
|
M3#1138Endocranial cast of the corresponding cranium of Nyctiellus lepidus Type: "3D_surfaces"doi: 10.18563/m3.sf.1138 state:published |
Download 3D surface file |
Cheiromeles torquatus AMNH M-247585 View specimen
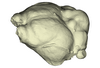
|
M3#1139Endocranial cast of the corresponding cranium of Cheiromeles torquatus Type: "3D_surfaces"doi: 10.18563/m3.sf.1139 state:published |
Download 3D surface file |
Miniopterus schreibersii UMMZ 156998 View specimen
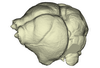
|
M3#1140Endocranial cast of the corresponding cranium of Miniopterus schreibersii Type: "3D_surfaces"doi: 10.18563/m3.sf.1140 state:published |
Download 3D surface file |
Kerivoula pellucida UMMZ 161396 View specimen

|
M3#1141Endocranial cast of the corresponding cranium of Kerivoula pellucida Type: "3D_surfaces"doi: 10.18563/m3.sf.1141 state:published |
Download 3D surface file |
Scotophilus kuhlii UMMZ 157013 View specimen
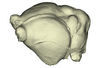
|
M3#1142Endocranial cast of the corresponding cranium of Scotophilus kuhlii Type: "3D_surfaces"doi: 10.18563/m3.sf.1142 state:published |
Download 3D surface file |
Rhinolophus luctus MNHN CG-2006-87 View specimen
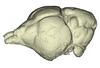
|
M3#1143Endocranial cast of the corresponding cranium of Rhinolophus luctus Type: "3D_surfaces"doi: 10.18563/m3.sf.1143 state:published |
Download 3D surface file |
Triaenops persicus AM RG-38552 View specimen
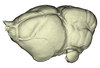
|
M3#1144Endocranial cast of the corresponding cranium of Triaenops persicus Type: "3D_surfaces"doi: 10.18563/m3.sf.1144 state:published |
Download 3D surface file |
Hipposideros armiger UM ZOOL-762-V View specimen
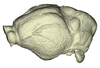
|
M3#1145Endocranial cast of the corresponding cranium of Hipposideros armiger Type: "3D_surfaces"doi: 10.18563/m3.sf.1145 state:published |
Download 3D surface file |
Lavia frons AM RG-12268 View specimen
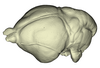
|
M3#1146Endocranial cast of the corresponding cranium of Lavia frons Type: "3D_surfaces"doi: 10.18563/m3.sf.1146 state:published |
Download 3D surface file |
Rhinopoma hardwickii AM RG-M31166 View specimen
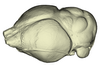
|
M3#1147Endocranial cast of the corresponding cranium of Rhinopoma hardwickii Type: "3D_surfaces"doi: 10.18563/m3.sf.1147 state:published |
Download 3D surface file |
Sphaerias blanfordi AMNH M-274330 View specimen
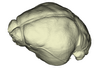
|
M3#1148Endocranial cast of the corresponding cranium of Sphaerias blanfordi Type: "3D_surfaces"doi: 10.18563/m3.sf.1148 state:published |
Download 3D surface file |
Rousettus aegyptiacus UMMZ 161026 View specimen
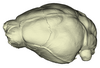
|
M3#1149Endocranial cast of the corresponding cranium of Rousettus aegyptiacus Type: "3D_surfaces"doi: 10.18563/m3.sf.1149 state:published |
Download 3D surface file |
Pteropus pumilus UMMZ 162253 View specimen
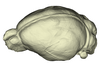
|
M3#1150Endocranial cast of the corresponding cranium of Pteropus pumilus Type: "3D_surfaces"doi: 10.18563/m3.sf.1150 state:published |
Download 3D surface file |
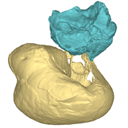
This contribution contains the 3D models described and figured in the following publication: Mourlam, M., Orliac, M. J. (2017), Protocetid (Cetacea, Artiodactyla) bullae and petrosals from the Middle Eocene locality of Kpogamé, Togo: new insights into the early history of cetacean hearing. Journal of Systematic Palaeontology https://doi.org/10.1080/14772019.2017.1328378
?Carolinacetus indet. UM KPG-M 164 View specimen
|
M3#132left petrosal of ?Carolinacetus sp. from the locality of Kpogamé, Togo Type: "3D_surfaces"doi: 10.18563/m3.sf.132 state:published |
Download 3D surface file |
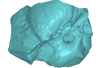
indet. indet. UM KPG-M 73 View specimen
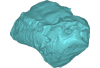
|
M3#133labelled surface of the left petrosal Type: "3D_surfaces"doi: 10.18563/m3.sf.133 state:published |
Download 3D surface file |

|
M3#134left bullaof Protocetidae indeterminate from Kpogamé, Togo Type: "3D_surfaces"doi: 10.18563/m3.sf.134 state:published |
Download 3D surface file |
|
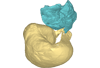
M3#135petrotympanic complex of Protocetidae indeterminate from Kpogamé, Togo Type: "3D_surfaces"doi: 10.18563/m3.sf.135 state:published |
Download 3D surface file |
?Carolinacetus indet. UM KPG-M 33 View specimen

|
M3#136left auditory bulla of a juvenile specimen of ?Carolinacetus sp. from Kpogamé, Togo Type: "3D_surfaces"doi: 10.18563/m3.sf.136 state:published |
Download 3D surface file |
Togocetus traversei UM KPG-M 80 View specimen

|
M3#137fragmentary right auditory bulla of Togocetus traversei from Kpogamé, Togo Type: "3D_surfaces"doi: 10.18563/m3.sf.137 state:published |
Download 3D surface file |

This contribution contains the 3D models of the bony labyrinths of two protocetid archaeocetes from the locality of Kpogamé, Togo, described and figured in the publication of Mourlam and Orliac (2017). https://doi.org/10.1016/j.cub.2017.04.061
?Carolinacetus indet. UM KPG-M 164 View specimen

|
M3#149bony labyrinth of ? Carolinacetus sp. from Kpogamé, Togo Type: "3D_surfaces"doi: 10.18563/m3.sf.149 state:published |
Download 3D surface file |
indet. indet. UM KPG-M 73 View specimen

|
M3#150bony labyrinth of Protocetidae indet. from Kpogamé, Togo Type: "3D_surfaces"doi: 10.18563/m3.sf.150 state:published |
Download 3D surface file |

This contribution contains the 3D model described and figured in the following publication: Hautier L, Sarr R, Lihoreau F, Tabuce R, Marwan Hameh P. 2014. First record of the family Protocetidae in the Lutetian of Senegal (West Africa). Palaeovertebrata 38(2)-e2
indet. indet. SN103 View specimen
|
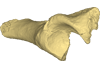
M3#5SN103, partial left innominate. Age and occurrence – Taïba Formation, Lutetian of the near Taïba Ndiaye, quarry of the Industries Chimiques du Sénégal (ICS) Type: "3D_surfaces"doi: 10.18563/m3.sf5 state:published |
Download 3D surface file |

The present 3D dataset contains 3D models of new material from the middle Eocene of the Upper Subathu Formation in the Kalakot area (India), documenting the anterior dentition of the raoellid Indohyus indirae. Raoellidae are closely related to stem cetaceans and bring crucial information to understand the earliest phase of land to water transition in Cetacea.
Indohyus indirae GU/RJ/31 View specimen

|
M3#1505Right i1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1505 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/32 View specimen

|
M3#1506Right i1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1506 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/16 View specimen

|
M3#1507Left I3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1507 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/23 View specimen

|
M3#1508Left I2 Type: "3D_surfaces"doi: 10.18563/m3.sf.1508 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/25 View specimen

|
M3#1509Left I1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1509 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/26 View specimen

|
M3#1510Right I3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1510 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/57 View specimen

|
M3#1511Left I1 Type: "3D_surfaces"doi: 10.18563/m3.sf.1511 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/61 View specimen

|
M3#1512Right upper canine Type: "3D_surfaces"doi: 10.18563/m3.sf.1512 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/63 View specimen

|
M3#1513Left upper canine Type: "3D_surfaces"doi: 10.18563/m3.sf.1513 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/74 View specimen

|
M3#1514Left upper canine Type: "3D_surfaces"doi: 10.18563/m3.sf.1514 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/439 View specimen

|
M3#1515Left upper canine Type: "3D_surfaces"doi: 10.18563/m3.sf.1515 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/457 View specimen

|
M3#1516Left upper canine Type: "3D_surfaces"doi: 10.18563/m3.sf.1516 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/846 View specimen

|
M3#1517Left upper canine Type: "3D_surfaces"doi: 10.18563/m3.sf.1517 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/822 View specimen
|
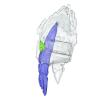
M3#1518Right fragmentary maxillary with decidual canine and I3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1518 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/824 View specimen

|
M3#1519Right fragmentary mandible with lower canine and small part of the i3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1519 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/838 View specimen

|
M3#1520Right fragmentary mandible with permanent i2, i3 and canine and small part of the root of the decidual i3 Type: "3D_surfaces"doi: 10.18563/m3.sf.1520 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/842 View specimen

|
M3#1521Left fragmentary mandible with decidual and permanent canine Type: "3D_surfaces"doi: 10.18563/m3.sf.1521 state:published |
Download 3D surface file |
Indohyus indirae GU/RJ/26,32,56,57,457,822,838,842 : Composite Anterior dentition View specimen

|
M3#15293D composite reconstruction of the anterior dentition of Indohyus indirae with GU/RJ/57 (I1), 56 (I2), 26 (I3), 457 (Upper canine), 822 (P1), 32 (i1), 838 (i2, i3 and lower canine) and 842 (p1) Type: "3D_surfaces"doi: 10.18563/m3.sf.1529 state:published |
Download 3D surface file |